Creating a virtual heart
To produce a piece of art you will first need a canvas
Patricia Martínez Díaz
27. September. 2021
Hey! Welcome back. It’s been almost a year since the beginning of my PhD and I would like to celebrate my first 12 months working as a researcher by sharing with you a little bit of my experience and some of the challenges that I have encountered while trying to generate a virtual heart and what do scientists mean when they talk about a digital twin.
The idea of using computer simulations to study the heart is not new. Almost sixty years ago (1962), Denis Noble used the mathematical model developed by Hodking and Huxley to describe the electrical activity of a cardiac cell [1]. From that moment on, researchers have been working to simulate not only single cardiac cells but also the entire organ. One of the most ambitious scientific projects of recent times is the development of a cardiac digital-twin: a virtual model of the heart that mimics patient’s individual anatomy and function (Figure 1). This multidisciplinary project seeks to replicate the complexity of the human heart with the use of computer simulations, so to help clinicians diagnose and plan therapy according to the specific needs of each patient, hence the importance of generating these virtual models. The development of a digital twin requires the integration of different levels of information: from single cell to tissue level, then from the organ level to finally the whole cardiovascular system. Although the development of the digital twin might not be an easy task it could for sure change the course of cardiology. That’s why efforts from the whole international cardiac modeling community must be joined up.

1. The Field of Computational Cardiology
In a very general way, the field of computational cardiology studies the heart from three different perspectives. Scientists can use computers to analyze for example the mechanical contraction of myocardial fibers to identify whether the heart has enough force to adequately pump blood. This branch is called computational biomechanics. Another approach is to study the blood movement inside the heart. A clear example is the use of simulations to identify the risk of stroke (when a blot clot clogs an artery) by analyzing blood flow patterns which can provide a hint to identify regions where flow is turbulent. This field is called computational fluid dynamics. Finally, researchers can also investigate the electrical activation of the heart to locate chaotic impulses that trigger an arrhythmia. The latter is called computational electrophysiology (see Figure 2). We have to keep in mind that during each beat, all the three mechanisms are active: biomechanics, fluid dynamics (also called hemodynamics) and electrophysiology.
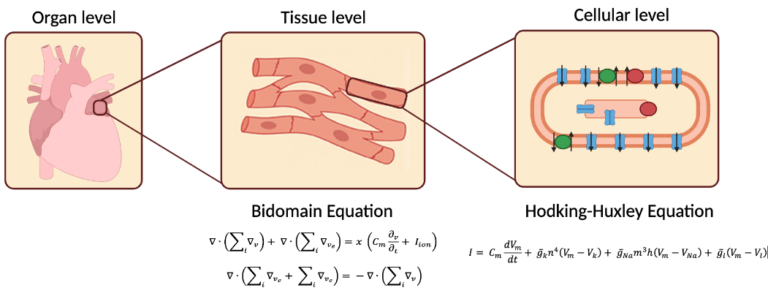
Ideally, a virtual heart would integrate the three aforementioned mechanisms. However, it is still possible to create a virtual heart that focuses specifically on only one of these aspects. For example, the use of computational electrophysiology for planning ablation therapies for patients with atrial fibrillation (AF), we are interested in the creation of a virtual heart in which we can simulate arrhythmias and perform simulated ablation lines. In the next section I am going to describe some of the steps needed to generate a virtual heart to study arrhythmias and help plan therapy.
2. First things first: the cardiac geometry
Irrespective of the computational model to be used, to generate our virtual heart we would first need to acquire the cardiac geometry of the patient. There are non-invasive methods such as Magnetic Resonance Imaging (MRI) and Computed Tomography (CT) scans which can provide detailed information of the heart anatomy. In addition, during a minimally-invasive procedure, such as an electrophysiological study, we can also get information about the geometry by using an electroanatomical mapping system (EAMS). All of these methods have advantages and limitations.
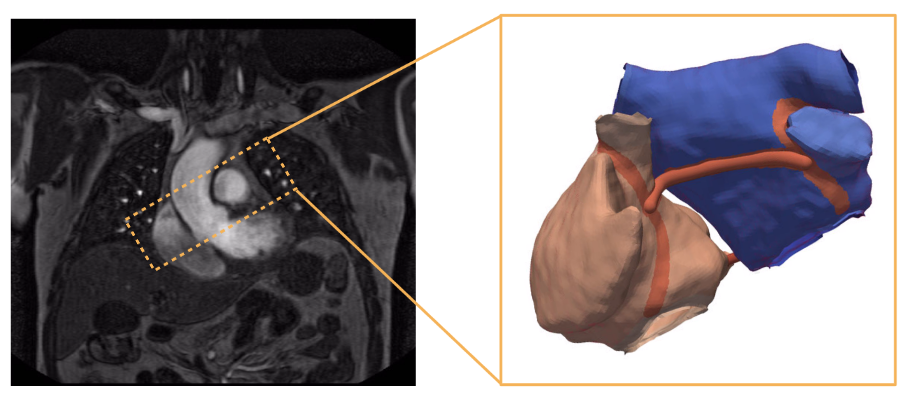
For example, non-invasive methods do not require the patient to enter the operating room. However, these studies can be costly and it can happen that only few patients get a MRI or CT scan. On the other hand, with the use of the EAMS we could not only get the geometry but we could also obtain information about the electrical properties of the heart which could be used later to tune our electrophysiological computer model. In the end, there is always a trade-off between advantages and disadvantages.
Acquiring the geometry is just the beginning of developing a virtual heart, however, we would also need to know more about the real anatomy of the patient. For example, some investigators have linked the presence of fibrosis in the atria to the occurrence of AF. So in order to create a personalized digital twin, it would be ideal not only to have the cardiac geometry but also the amount of fibrosis and also its distribution throughout the atria. The challenge now is to identify methods that are capable of capturing the specific distribution of fibrosis for each patient, and to translate this data into the computer model. One of them is the use late gadolinium enhancement (LGE) MRI to detect fibrosis. An additional approach used to assess the location of fibrotic tissue is through the use of mapping catheters inside the atria and to measure the voltage. Finally, computer modelers could incorporate these data into the virtual heart.
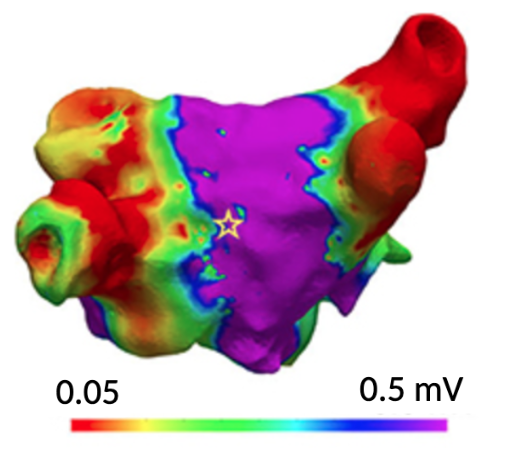
Take a look at the following image courtesy of my colleague Deborah Nairn. What you are looking at is a digital version of the left atrium, where the colors represent voltage levels measured with the help of a catheter inserted into the heart.
Of course, having a geometry is only the first step for generating a virtual model of the heart. Let’s put it this way, this is the canvas on which we will begin to paint our work. In the following blogs I will tell you a bit more about the additional steps needed to create a digital twin of the heart. If you are interested in these topics on computational cardiology you can follow us on all our social media. See you next time and stay well.
3. References
- Noble, D., (1962), A modification of the Hodgkin—Huxley equations applicable to Purkinje fibre action and pacemaker potentials. The Journal of Physiology, 160 doi: 10.1113/jphysiol.1962.sp006849
- Trayanova, N., (2011) Whole-Heart Modeling: Applications to Cardiac Electrophysiology and Electromechanics. Alternans and Arrhythmias: From Cells to the Heart [Circ Res. 2011;108:98–112]
- Nairn, D., (2021) Comparison of Unipolar and Bipolar Voltage Mapping for Localization of Left Atrial Arrhythmogenic Substrate in Patients With Atrial Fibrillation. Front. Physiol., 26 November 2020 10.3389/fphys.2020.575846
Follow us in our social media accounts to stay tuned, and contact to personalizeAF@itaca.upv.es if you want to know more about our project!